Graph of compute time as function of dimension
We can use the functions
compare_eta_comptime_across_nvars() and
plot_eta_comptime() to investigate computation time of the
algorithm for varying dimension of the parameter vector
.
This can also be used to investigate computation time for different
sample methods, different tuning parameters, or whatever the user might
be interested in.
The basic usage of the function can be used to investigate the computational gain for high dimensions of the CGGibbs sampler compared to the naive approach of doing computation for each coordinate update. For this purpose, the function call is:
res <- compare_eta_comptime_across_nvars(
n_vars = c(2, seq(from = 50, to = 500, by = 50)),
n_samples = 1,
burnin = 0,
w = 0.5)The results can be plotted by simply calling
plot_eta_comptime(res)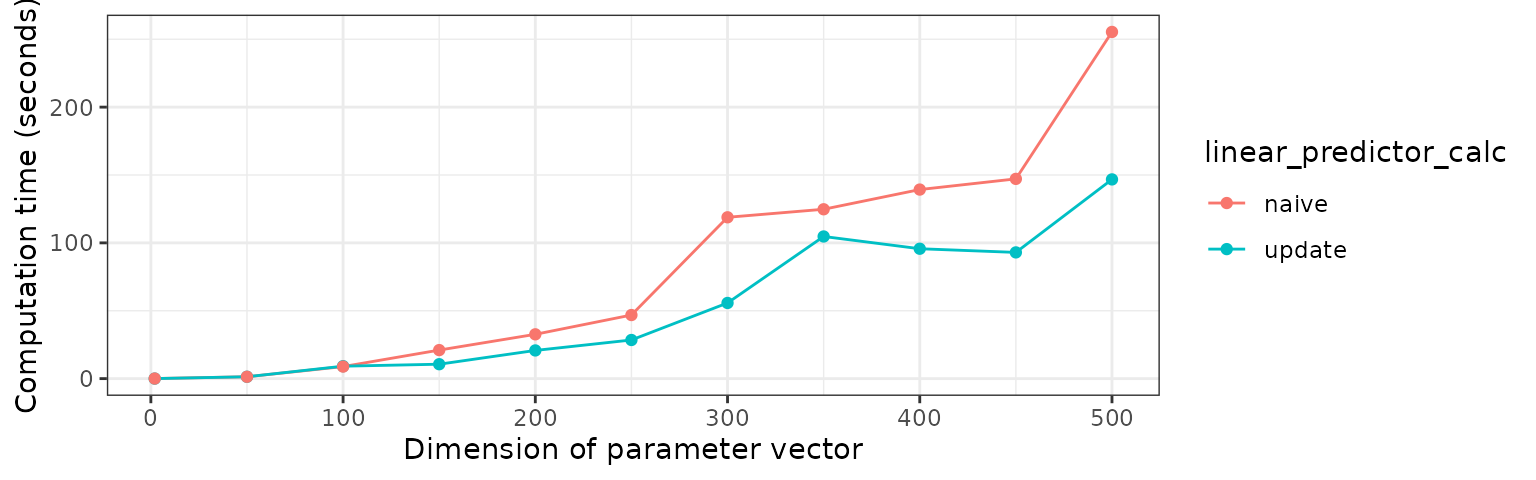
Runtime across dimensions for different sampling methods
The user can easily just iterate the function across different
sampling procedures or tuning parameters for a single sample procedure,
bind the results, and then use the facet_by argument in
plot_eta_comptime().
An example of this could look like: > We won’t run this code for this vignette to reduce rendering time, but it provides an idea to what is possible using the functionalities.
ws <- c(0.5, 5, 20, 100)
res <- lapply(ws, function(w) {
compare_eta_comptime_across_nvars(
w = w,
n_vars = c(2, seq(from = 10, to = 50, by = 10)),
n_samples = 50,
burnin = 1,
parallelise = TRUE)
}) %>%
dplyr::bind_rows()
plot_eta_comptime(res, facet_by = "w")